Identification and transcriptome analysis of the R2R3-MYB gene family in Haloxylon ammodendron
DOI:
https://doi.org/10.55779/nsb15411649Keywords:
Haloxylon ammodendron, R2R3-MYB family, plant stress resistance, RNA-seq, bioinformationAbstract
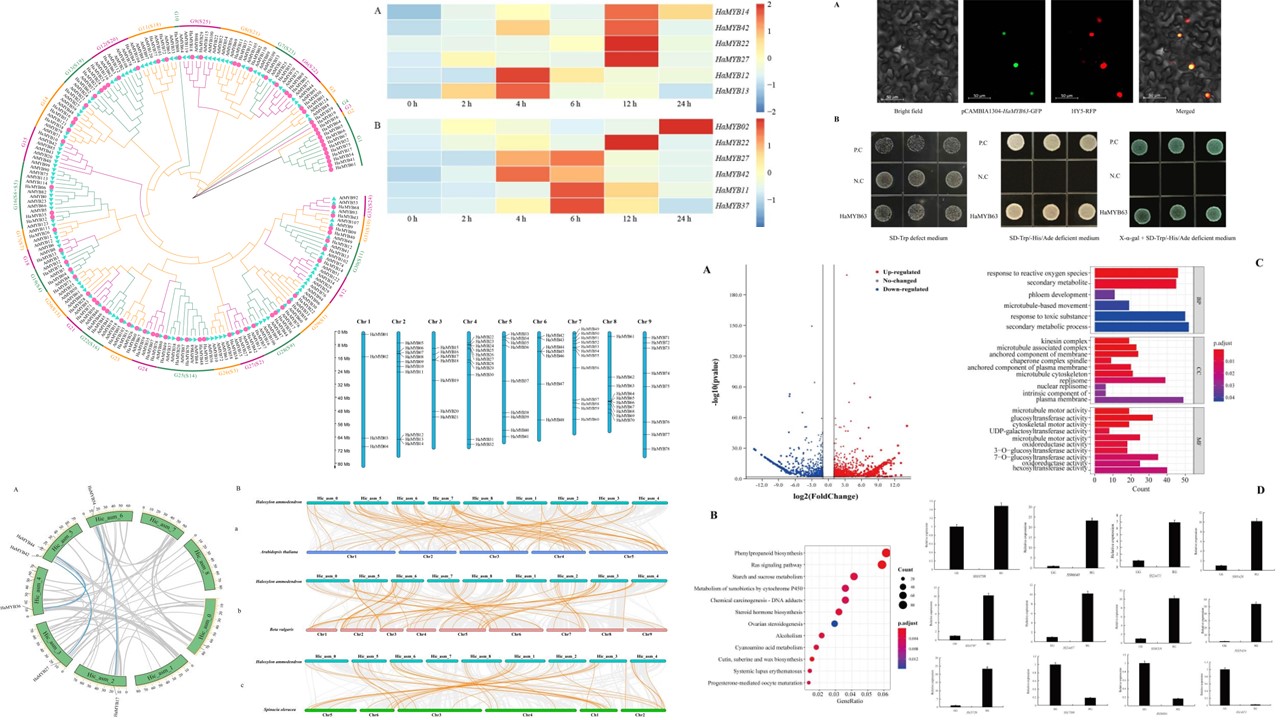
The MYB transcription factor family is widespread in plants and plays an important role in plant growth and development as well as in plant responses to stress. The MYB transcription factor family has been identified in a variety of organisms; however, it has not been identified and analysed in the desert plant Haloxylon ammodendron. In this study, R2R3-MYB genes were identified and analysed using a bioinformatic approach. A total of 78 R2R3-MYB genes were identified and named according to their position on the chromosome. The R2R3-MYB genes were unevenly distributed on nine chromosomes. Phylogenetic analysis showed that the HaMYB genes were all divided into 31 subfamilies. Covariance analysis revealed the presence of three pairs of fragmentary duplicated genes in H. ammodendron (HaMYB54 and HaMYB17, HaMYB44 and HaMYB36, HaMYB42 and HaMYB27). Gene structure and conserved structural domain analysis revealed different subgroups with different orders of magnitude of variation in gene structures and conserved structural domains. Analysis of cis-elements showed that the cis-acting elements of HaMYBs were mainly associated with hormone and abiotic stress responses. Real-time quantitative PCR was used to detect the expression levels of HaR2R3-MYB genes, and six HaR2R3-MYB genes were found to respond to salt stress and six HaR2R3-MYB genes to drought stress, with HaMYB22 and HaMYB27 showing upregulated expression under both stresses. Transcriptome analysis showed that HaMYB63 was significantly differentially expressed in the assimilated branches of H. ammodendron, and the subcellular localization of this protein showed that it was located in the nucleus and had transcriptional self-activating activity. These results provide a theoretical basis for further studies on the functions of the R2R3-MYB gene family and the molecular mechanisms of resistance in H. ammodendron.
Metrics
References
Abubakar AS, Feng X, Gao G, Yu C, Chen J, Chen K, ... Zhu A (2022). Genome wide characterization of R2R3 MYB transcription factor from Apocynum venetum revealed potential stress tolerance and flavonoid biosynthesis genes. Genomics 114(2):110275. https://doi.org/10.1016/j.ygeno.2022.110275
Ambawat S, Sharma P, Yadav NR, Yadav RC (2013). MYB transcription factor genes as regulators for plant responses: an overview. Physiology and Molecular Biology of Plants 19:307-321. https://doi.org/10.1007/s12298-013-0179-1
Ampomah-Dwamena C, Thrimawithana AH, Dejnoprat S, Lewis D, Espley RV, Allan AC (2019). A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytologist 221(1):309-325. https://doi.org/10.1111/nph.15362
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, De Castro E, ... Stockinger H (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research 40(W1):W597-W603. https://doi.org/10.1093/nar/gks400
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37:W202-W208. https://doi.org/10.1093/nar/gkp335
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant 13(8):1194-1202. https://doi.org/10.1016/j.molp.2020.06.009
Cui J, Qiu T, Li L, Cui S (2023). De novo full-length transcriptome analysis of two ecotypes of Phragmites australis (swamp reed and dune reed) provides new insights into the transcriptomic complexity of dune reed and its long-term adaptation to desert environments. BMC Genomics 24(1):1-23. https://doi.org/10.1186/s12864-023-09271-y
Du H, Feng BR, Yang SS, Huang YB, Tang YX (2012). The R2R3-MYB transcription factor gene family in maize. PloS One 7(6):e37463. https://doi.org/10.1371/journal.pone.0037463
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010). MYB transcription factors in Arabidopsis. Trends in Plant Science 15(10):573-581. https://doi.org/10.1016/j.tplants.2010.06.005
Feng G, Burleigh JG, Braun EL, Mei W, Barbazuk WB (2017). Evolution of the 3R-MYB gene family in plants. Genome Biology and Evolution 9(4):1013-1029. https://doi.org/10.1093/gbe/evx056
Gao F, Zhao HX, Yao HP, Li CL, Chen H, Wang AH, ... Wu Q (2016). Identification, isolation and expression analysis of eight stress-related R2R3-MYB genes in tartary buckwheat (Fagopyrum tataricum). Plant Cell Reports 35:1385-1396. https://doi.org/10.1007/s00299-016-1971-5
Gao H, Lü X, Ren W, Sun Y, Zhao Q, Wang G, ... Zhang J (2020). HaASR1 gene cloned from a desert shrub, Haloxylon ammodendron, confers drought tolerance in transgenic Arabidopsis thaliana. Environmental and Experimental Botany 180:104251. https://doi.org/10.1016/j.envexpbot.2020.104251
He AL, Niu SQ, Zhao Q, Li YS, Gou JY, Gao HJ, ... Zhang JL (2018). Induced salt tolerance of perennial ryegrass by a novel bacterium strain from the rhizosphere of a desert shrub Haloxylon ammodendron. International Journal of Molecular Sciences 19(2):469. https://doi.org/10.3390/ijms19020469
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Research 35:W585-W587. https://doi.org/10.1093/nar/gkm259
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:1-19. https://doi.org/10.1186/1471-2164-13-544
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology 37(8):907-915. https://doi.org/10.1038/s41587-019-0201-4
Klempnauer KH, Gonda TJ, Bishop JM (1982). Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene. Cell 31(2):453-463. https://doi.org/10.1016/0092-8674(82)90138-6
Li J, Chang H, Liu T, Zhang C (2019). The potential geographical distribution of Haloxylon across Central Asia under climate change in the 21st century. Agricultural and Forest Meteorology 275:243-254. https://doi.org/10.1016/j.agrformet.2019.05.027
Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402-408. https://doi.org/10.1006/meth.2001.1262
Lotkowska ME, Tohge T, Fernie AR, Xue GP, Balazadeh S, Mueller-Roeber B (2015). The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant physiology 169(3):1862-1880. https://doi.org/10.1104/pp.15.00605
Love MI, Huber W, Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15(12):1-21. https://doi.org/10.1186/s13059-014-0550-8
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, ... Marchler-Bauer A (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Research 48(D1):D265-D268. https://doi.org/10.1093/nar/gkz991
Lü XP, Gao HJ, Zhang L, Wang YP, Shao KZ, Zhao Q, Zhang JL (2019). Dynamic responses of Haloxylon ammodendron to various degrees of simulated drought stress. Plant Physiology and Biochemistry 139:121-131. https://doi.org/10.1016/j.plaphy.2019.03.019
Lü XP, Shao KZ, Xu JY, Li JL, Ren W, Chen J, ... Zhang JL (2022). A heat shock transcription factor gene (HaHSFA1) from a desert shrub, Haloxylon ammodendron, elevates salt tolerance in Arabidopsis thaliana. Environmental and Experimental Botany 201:104954. https://doi.org/10.1016/j.envexpbot.2022.104954
Ma Q, Wang X, Chen F, Wei L, Zhang D, Jin H (2021). Carbon sequestration of sand-fixing plantation of Haloxylon ammodendron in Shiyang River Basin: storage, rate and potential. Global Ecology and Conservation 28:e01607. https://doi.org/10.1016/j.gecco.2021.e01607
Manna M, Thakur T, Chirom O, Mandlik R, Deshmukh R, Salvi P (2021). Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiologia Plantarum 172(2):847-868. https://doi.org/10.1111/ppl.13268
Matus JT, Aquea F, Arce-Johnson P (2008). Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology 8:1-15. https://doi.org/10.1186/1471-2229-8-83
Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. The EMBO Journal 6(12):3553-3558. https://doi.org/10.1002/j.1460-2075.1987.tb02684.x
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology 33(3):290-295. https://doi.org/10.1038/nbt.3122
Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD (2018). HMMER web server: 2018 update. Nucleic Acids Research 46(W1):W200-W204. https://doi.org/10.1093/nar/gky448
Romani F, Moreno JE (2021). Molecular mechanisms involved in functional macroevolution of plant transcription factors. New Phytologist 230(4):1345-1353. https://doi.org/10.1111/nph.17161
Rusconi F, Simeoni F, Francia P, Cominelli E, Conti L, Riboni M, ... Galbiati M (2013). The Arabidopsis thaliana MYB60 promoter provides a tool for the spatio-temporal control of gene expression in stomatal guard cells. Journal of Experimental Botany 64(11):3361-3371. https://doi.org/10.1093/jxb/ert180
Salih H, Bai W, Zhao M, Liang Y, Yang R, Zhang D, Li X (2023). Genome-wide characterization and expression analysis of transcription factor families in desert moss Syntrichia caninervis under abiotic stresses. International Journal of Molecular Sciences 24(7):6137. https://doi.org/10.3390/ijms24076137
Shen XJ, Wang YY, Zhang YX, Guo W, Jiao YQ, Zhou XA (2018). Overexpression of the wild soybean R2R3-MYB transcription factor GsMYB15 enhances resistance to salt stress and Helicoverpa armigera in transgenic Arabidopsis. International Journal of Molecular Sciences 19(12):3958. https://doi.org/10.3390/ijms19123958
Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. The Plant Cell 19(8):2440-2453. https://doi.org/10.1105/tpc.107.050963
Shriti S, Paul S, Das S (2023). Overexpression of CaMYB78 transcription factor enhances resistance response in chickpea against Fusarium oxysporum and negatively regulates anthocyanin biosynthetic pathway. Protoplasma 260(2):589-605. https://doi.org/10.1007/s00709-022-01797-4
Song J, Feng G, Tian CY, Zhang FS (2006). Osmotic adjustment traits of Suaeda physophora, Haloxylon ammodendron and Haloxylon persicum in field or controlled conditions. Plant Science 170(1):113-119. https://doi.org/10.1016/j.plantsci.2005.08.004
Stracke R, Holtgräwe D, Schneider J, Pucker B, Rosleff Sörensen T, Weisshaar B (2014). Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biology 14(1):1-17. https://doi.org/10.1186/s12870-014-0249-8
Stracke R, Werber M, Weisshaar B (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Current Opinion in Plant Biology 4(5):447-456. https://doi.org/10.1016/S1369-5266(00)00199-0
Subramanian B, Gao S, Lercher MJ, Hu S, Chen WH (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Research 47(W1):W270-W275. https://doi.org/10.1093/nar/gkz357
Sun J, Xu J, Qu W, Han X, Qiu C, Gai Z, ... Li Z (2023). Genome-wide analysis of R2R3-MYB transcription factors reveals their differential responses to drought stress and ABA treatment in desert poplar (Populus euphratica). Gene 855:147124. https://doi.org/10.1016/j.gene.2022.147124
Sun W, Ma Z, Chen H, Liu M (2019). MYB gene family in potato (Solanum tuberosum L.): genome-wide identification of hormone-responsive reveals their potential functions in growth and development. International Journal of Molecular Sciences 20(19):4847. https://doi.org/10.3390/ijms20194847
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30(12):2725-2729. https://doi.org/10.1093/molbev/mst197
Voorrips R (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93(1):77-78. https://doi.org/10.1093/jhered/93.1.77
Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, ... Paterson AH (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research 40(7):e49-e49. https://doi.org/10.1093/nar/gkr1293
Wang Z, Ni L, Hua J, Liu L, Yin Y, Li H, Gu C (2021). Transcriptome analysis reveals regulatory framework for salt and drought tolerance in Hibiscus hamabo siebold & zuccarini. Forests 12(4):454. https://doi.org/10.3390/f12040454
Wyrzykowska A, Bielewicz D, Plewka P, Sołtys-Kalina D, Wasilewicz-Flis I, Marczewski W, ... Szweykowska-Kulinska Z (2022). The MYB33, MYB65, and MYB101 transcription factors affect Arabidopsis and potato responses to drought by regulating the ABA signaling pathway. Physiologia Plantarum 174(5):e13775. https://doi.org/10.1111/ppl.13775
Yang B, Li Y, Song Y, Wang X, Guo Q, Zhou L, ... Zhang C (2022). The R2R3-MYB transcription factor VcMYB4a inhibits lignin biosynthesis in blueberry (Vaccinium corymbosum). Tree Genetics & Genomes 18(3):27. https://doi.org/10.1007/s11295-022-01560-z
Yao T, Zhang J, Xie M, Yuan G, Tschaplinski TJ, Muchero W, Chen JG (2021). Transcriptional regulation of drought response in Arabidopsis and woody plants. Frontiers in Plant Science 11:572137. https://doi.org/10.3389/fpls.2020.572137
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:1-11. https://doi.org/10.1186/1471-2105-13-134
Zhang S, Zhao Q, Zeng D, Xu J, Zhou H, Wang F, ... Li Y (2019). RhMYB108, an R2R3-MYB transcription factor, is involved in ethylene-and JA-induced petal senescence in rose plants. Horticulture Research 6:131. https://doi.org/10.1038/s41438-019-0221-8
Zhang, K, Su Y, Wang T, Liu T (2016). Soil properties and herbaceous characteristics in an age sequence of Haloxylon ammodendron plantations in an oasis-desert ecotone of northwestern China. Journal of Arid Land 8:960-972. https://doi.org/10.1007/s40333-016-0096-6
Zhao K, Cheng Z, Guo Q, Yao W, Liu H, Zhou B, Jiang T (2020). Characterization of the poplar R2R3-MYB gene family and over-expression of PsnMYB108 confers salt tolerance in transgenic tobacco. Frontiers in Plant Science 11:571881. https://doi.org/10.3389/fpls.2020.571881
Zhou Z, Wei X, Lan H (2023). CgMYB1, an R2R3-MYB transcription factor, can alleviate abiotic stress in an annual halophyte Chenopodium glaucum. Plant Physiology and Biochemistry 196:484-496. https://doi.org/10.1016/j.plaphy.2023.01.055
Zhu L, Guan Y, Zhang Z, Song A, Chen S, Jiang J, Chen F (2020). CmMYB8 encodes an R2R3 MYB transcription factor which represses lignin and flavonoid synthesis in chrysanthemum. Plant Physiology and Biochemistry 149:217-224. https://doi.org/10.1016/j.plaphy.2020.02.010
Zhu Y, Hu X, Wang P, Wang H, Ge X, Li F, Hou Y (2022). GhODO1, an R2R3-type MYB transcription factor, positively regulates cotton resistance to Verticillium dahliae via the lignin biosynthesis and jasmonic acid signaling pathway. International Journal of Biological Macromolecules 201:580-591. https://doi.org/10.1016/j.ijbiomac.2022.01.120
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Guohui ZHOU, Yanping REN, Li MA, Cong CHENG, Bo WANG, Zhengpei YAO, Shanshan LI, Meini TAO, Yue ZHAO, Zhiqiang LI, Hua ZHANG

This work is licensed under a Creative Commons Attribution 4.0 International License.
Papers published in Notulae Scientia Biologicae are Open-Access, distributed under the terms and conditions of the Creative Commons Attribution License.
© Articles by the authors; licensee SMTCT, Cluj-Napoca, Romania. The journal allows the author(s) to hold the copyright/to retain publishing rights without restriction.
License:
Open Access Journal - the journal offers free, immediate, and unrestricted access to peer-reviewed research and scholarly work, due SMTCT supports to increase the visibility, accessibility and reputation of the researchers, regardless of geography and their budgets. Users are allowed to read, download, copy, distribute, print, search, or link to the full texts of the articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author.













.png)















